Do you know that you can have ggplot in BBC R graphics cookbook? This is an attempt to reproduce https://bbc.github.io/rcookbook/ in python
Difference between plotnine and ggplot
99% of them are the same, except that in python you have to wrap column names in '', otherwise it will be treated as variable and caused error. Most of the time you just need to wrap a '' or replaced with _ depends on the function.
I tried to produce the same chart with plotnine and altair, and hopefully you will see their difference. plotnine covers 99% of ggplot2, so if you are coming from R, just go ahead with plotnine! altair is another interesting visualization library that base on vega-lite, therefore it can be integrated with website easily. In addition, it can also produce interactive chart with very simple function, which is a big plus!
Setup
# collapse-hide # !pip install plotnine[all] # !pip install altair # !pip install gapminder from gapminder import gapminderfrom plotnine.data import mtcarsfrom plotnine import * from plotnine import ggplot, geom_point, aes, stat_smooth, facet_wrap, geom_linefrom plotnine import ggplot # https://plotnine.readthedocs.io/en/stable/ import altair as altimport pandas as pdimport plotnine% matplotlib inline
Collecting plotnine[all]
Using cached https://files.pythonhosted.org/packages/19/da/4d2f68e7436e76a3c26ccd804e1bfc5c58fca7a6cba06c71bab68b25e825/plotnine-0.6.0-py3-none-any.whl
Collecting descartes>=1.1.0 (from plotnine[all])
Using cached https://files.pythonhosted.org/packages/e5/b6/1ed2eb03989ae574584664985367ba70cd9cf8b32ee8cad0e8aaeac819f3/descartes-1.1.0-py3-none-any.whl
Requirement already satisfied: numpy>=1.16.0 in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from plotnine[all]) (1.16.5)
Requirement already satisfied: matplotlib>=3.1.1 in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from plotnine[all]) (3.1.1)
Requirement already satisfied: statsmodels>=0.9.0 in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from plotnine[all]) (0.10.1)
Requirement already satisfied: pandas>=0.25.0 in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from plotnine[all]) (1.0.3)
Requirement already satisfied: scipy>=1.2.0 in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from plotnine[all]) (1.3.1)
Requirement already satisfied: patsy>=0.4.1 in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from plotnine[all]) (0.5.1)
Collecting mizani>=0.6.0 (from plotnine[all])
Using cached https://files.pythonhosted.org/packages/e3/76/7a2c9094547ee592f9f43f651ab824aa6599af5e1456250c3f4cc162aece/mizani-0.6.0-py2.py3-none-any.whl
Requirement already satisfied: scikit-learn; extra == "all" in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from plotnine[all]) (0.22.1)
Collecting scikit-misc; extra == "all" (from plotnine[all])
Using cached https://files.pythonhosted.org/packages/94/4c/e6c3ba02dc66278317778b5c5df7b372c6c5313fce43615a7ce7fc0b34b8/scikit_misc-0.1.1-cp37-cp37m-win_amd64.whl
Requirement already satisfied: cycler>=0.10 in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from matplotlib>=3.1.1->plotnine[all]) (0.10.0)
Requirement already satisfied: kiwisolver>=1.0.1 in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from matplotlib>=3.1.1->plotnine[all]) (1.1.0)
Requirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from matplotlib>=3.1.1->plotnine[all]) (2.4.2)
Requirement already satisfied: python-dateutil>=2.1 in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from matplotlib>=3.1.1->plotnine[all]) (2.8.0)
Requirement already satisfied: pytz>=2017.2 in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from pandas>=0.25.0->plotnine[all]) (2019.3)
Requirement already satisfied: six in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from patsy>=0.4.1->plotnine[all]) (1.12.0)
Requirement already satisfied: palettable in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from mizani>=0.6.0->plotnine[all]) (3.3.0)
Requirement already satisfied: joblib>=0.11 in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from scikit-learn; extra == "all"->plotnine[all]) (0.13.2)
Requirement already satisfied: setuptools in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from kiwisolver>=1.0.1->matplotlib>=3.1.1->plotnine[all]) (41.4.0)
Installing collected packages: descartes, mizani, scikit-misc, plotnine
Successfully installed descartes-1.1.0 mizani-0.6.0 plotnine-0.6.0 scikit-misc-0.1.1
Requirement already satisfied: altair in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (4.1.0)
Requirement already satisfied: jinja2 in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from altair) (2.10.3)
Requirement already satisfied: jsonschema in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from altair) (3.0.2)
Requirement already satisfied: entrypoints in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from altair) (0.3)
Requirement already satisfied: numpy in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from altair) (1.16.5)
Requirement already satisfied: toolz in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from altair) (0.10.0)
Requirement already satisfied: pandas>=0.18 in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from altair) (1.0.3)
Requirement already satisfied: MarkupSafe>=0.23 in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from jinja2->altair) (1.1.1)
Requirement already satisfied: pyrsistent>=0.14.0 in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from jsonschema->altair) (0.15.4)
Requirement already satisfied: setuptools in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from jsonschema->altair) (41.4.0)
Requirement already satisfied: six>=1.11.0 in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from jsonschema->altair) (1.12.0)
Requirement already satisfied: attrs>=17.4.0 in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from jsonschema->altair) (19.2.0)
Requirement already satisfied: pytz>=2017.2 in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from pandas>=0.18->altair) (2019.3)
Requirement already satisfied: python-dateutil>=2.6.1 in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from pandas>=0.18->altair) (2.8.0)
Collecting gapminder
Downloading https://files.pythonhosted.org/packages/85/83/57293b277ac2990ea1d3d0439183da8a3466be58174f822c69b02e584863/gapminder-0.1-py3-none-any.whl
Requirement already satisfied: pandas in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from gapminder) (1.0.3)
Requirement already satisfied: numpy>=1.13.3 in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from pandas->gapminder) (1.16.5)
Requirement already satisfied: pytz>=2017.2 in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from pandas->gapminder) (2019.3)
Requirement already satisfied: python-dateutil>=2.6.1 in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from pandas->gapminder) (2.8.0)
Requirement already satisfied: six>=1.5 in c:\users\channo.oocldm\appdata\local\continuum\anaconda3\lib\site-packages (from python-dateutil>=2.6.1->pandas->gapminder) (1.12.0)
Installing collected packages: gapminder
Successfully installed gapminder-0.1
print (f'altair version: { alt. __version__} ' )print (f'plotnine version: { plotnine. __version__} ' )print (f'pandas version: { pd. __version__} ' )
altair version: 4.1.0
plotnine version: 0.6.0
pandas version: 1.0.3
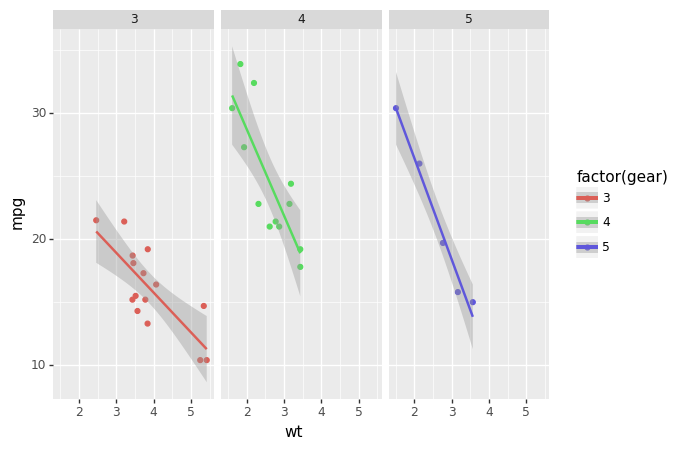
Plotnine Example
'wt' , 'mpg' , color= 'factor(gear)' ))+ geom_point()+ stat_smooth(method= 'lm' )+ facet_wrap('~gear' ))
Make a Line Chart
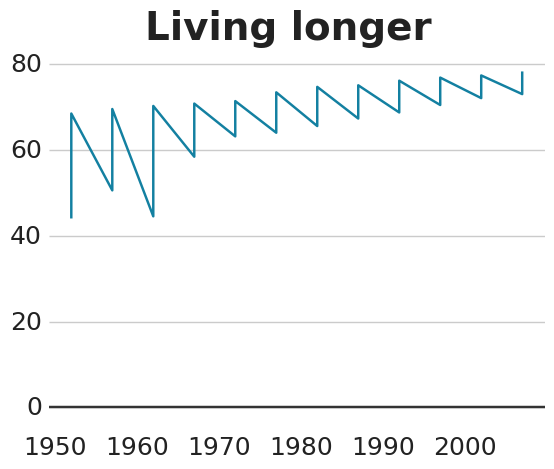
ggplot
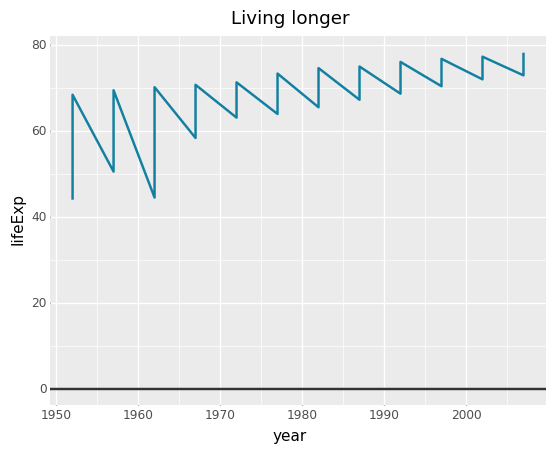
<- gapminder %>% filter (country == "Malawi" ) #Make plot <- ggplot (line_df, aes (x = year, y = lifeExp)) + geom_line (colour = "#1380A1" , size = 1 ) + geom_hline (yintercept = 0 , size = 1 , colour= "#333333" ) + bbc_style () + labs (title= "Living longer" ,subtitle = "Life expectancy in Malawi 1952-2007" )
#hide = gapminder.query(" country == 'Malawi' " )
plotnine
= 'year' , y= 'lifeExp' )) + = '#1380A1' , size= 1 ) + = 0 , size = 1 , colour= '#333333' ) + = 'Living longer' ,= 'Life expectancy in Malawi 1952-2007' )
## altair = (alt.Chart(line_df).mark_line().encode(= 'year' ,= 'lifeExp' )= {'text' : 'Living Longer' ,'subtitle' : 'Life expectancy in Malawi 1952-2007' })# hline = overlay = pd.DataFrame({'y' : [0 ]})= alt.Chart(overlay).mark_rule(color= '#333333' , strokeWidth= 3 ).encode(y= 'y:Q' )+ hline
The BBC style
function () <- "Helvetica" :: theme (plot.title = ggplot2:: element_text (family = font, size = 28 , face = "bold" , color = "#222222" ), plot.subtitle = ggplot2:: element_text (family = font, size = 22 , margin = ggplot2:: margin (9 , 0 , 9 , 0 )), plot.caption = ggplot2:: element_blank (), legend.position = "top" , legend.text.align = 0 , legend.background = ggplot2:: element_blank (), legend.title = ggplot2:: element_blank (), legend.key = ggplot2:: element_blank (), legend.text = ggplot2:: element_text (family = font, size = 18 , color = "#222222" ), axis.title = ggplot2:: element_blank (), axis.text = ggplot2:: element_text (family = font, size = 18 , color = "#222222" ), axis.text.x = ggplot2:: element_text (margin = ggplot2:: margin (5 , b = 10 )), axis.ticks = ggplot2:: element_blank (), axis.line = ggplot2:: element_blank (), panel.grid.minor = ggplot2:: element_blank (), panel.grid.major.y = ggplot2:: element_line (color = "#cbcbcb" ), panel.grid.major.x = ggplot2:: element_blank (), panel.background = ggplot2:: element_blank (), strip.background = ggplot2:: element_rect (fill = "white" ), strip.text = ggplot2:: element_text (size = 22 , hjust = 0 ))< environment: namespace: bbplot>
NameError: name 'legend_text_align' is not defined
def bbc_style():= "Helvetica" = theme(plot_title= element_text(family= font,= 28 , face= "bold" , color= "#222222" ),# plot_subtitle=element_text(family=font, # size=22, plot_margin=(9, 0, 9, 0)), plot_caption=element_blank(), = "top" , legend_title_align= 0 , legend_background= element_blank(),= element_blank(), legend_key= element_blank(),= element_text(family= font, size= 18 ,= "#222222" ), axis_title= element_blank(),= element_text(family= font, size= 18 ,= "#222222" ),= element_text(margin= {'t' : 5 , 'b' : 10 }),= element_blank(),= element_blank(), panel_grid_minor= element_blank(),= element_line(color= "#cbcbcb" ),= element_blank(), panel_background= element_blank(),= element_rect(fill= "white" ),= element_text(size= 22 , hjust= 0 )return t
= "Helvetica" = element_text(family= font,= 28 , face= "bold" , color= "#222222" ),# plot_subtitle=element_text(family=font, # size=22, plot_margin=(9, 0, 9, 0)), plot_caption=element_blank(), = "top" , legend_title_align= 0 , legend_background= element_blank(),= element_blank(), legend_key= element_blank(),= element_text(family= font, size= 18 ,= "#222222" ), axis_title= element_blank(),= element_text(family= font, size= 18 ,= "#222222" ),= element_text(margin= {'t' : 5 , 'b' : 10 }),= element_blank(),= element_blank(), panel_grid_minor= element_blank(),= element_line(color= "#cbcbcb" ),= element_blank(), panel_background= element_blank(),= element_rect(fill= "white" ),= element_text(size= 22 , hjust= 0 )
<plotnine.themes.theme.theme at 0x163f0ca1508>
The finalise_plot() function does more than just save out your chart, it also left-aligns the title and subtitle as is standard for BBC graphics, adds a footer with the logo on the right side and lets you input source text on the left side.
altair
## altair = (alt.Chart(line_df).mark_line().encode(= 'year' ,= 'lifeExp' )= {'text' : 'Living Longer' ,'subtitle' : 'Life expectancy in China 1952-2007' })# hline = overlay = pd.DataFrame({'lifeExp' : [0 ]})= alt.Chart(overlay).mark_rule(color= '#333333' , strokeWidth= 3 ).encode(y= 'lifeExp:Q' )+ hline
Make a multiple line chart
# hide # Prepare data = gapminder.query('country == "China" | country =="United States" ' )
ggplot
#Prepare data <- gapminder %>% filter (country == "China" | country == "United States" ) #Make plot <- ggplot (multiple_line_df, aes (x = year, y = lifeExp, colour = country)) + geom_line (size = 1 ) + geom_hline (yintercept = 0 , size = 1 , colour= "#333333" ) + scale_colour_manual (values = c ("#FAAB18" , "#1380A1" )) + bbc_style () + labs (title= "Living longer" ,subtitle = "Life expectancy in China and the US" )
plotnine
# Make plot = (= 'year' , y= 'lifeExp' , colour= 'country' )) + = "#1380A1" , size= 1 ) + = 0 , size= 1 , color= "#333333" ) + = ["#FAAB18" , "#1380A1" ]) + + = "Living longer" ,= "Life expectancy in China 1952-2007" ))
findfont: Font family ['Helvetica'] not found. Falling back to DejaVu Sans.
findfont: Font family ['Helvetica'] not found. Falling back to DejaVu Sans.
altair
= (alt.Chart(multiline_df).mark_line().encode(= 'year' ,= 'lifeExp' ,= 'country' )= {'text' : 'Living Longer' ,'subtitle' : 'Life expectancy in China 1952-2007' })# hline = overlay = pd.DataFrame({'lifeExp' : [0 ]})= alt.Chart(overlay).mark_rule(color= '#333333' , strokeWidth= 3 ).encode(y= 'lifeExp:Q' )+ hline
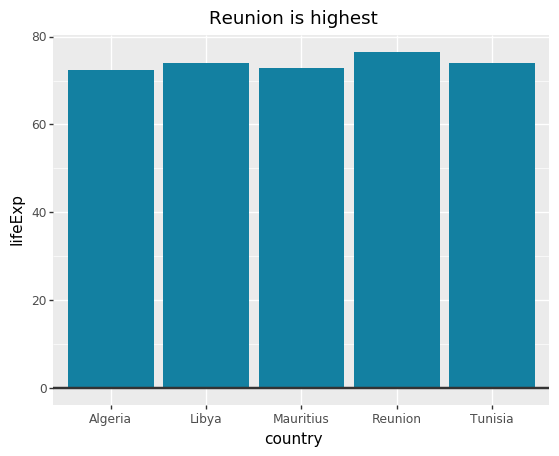
ggplot
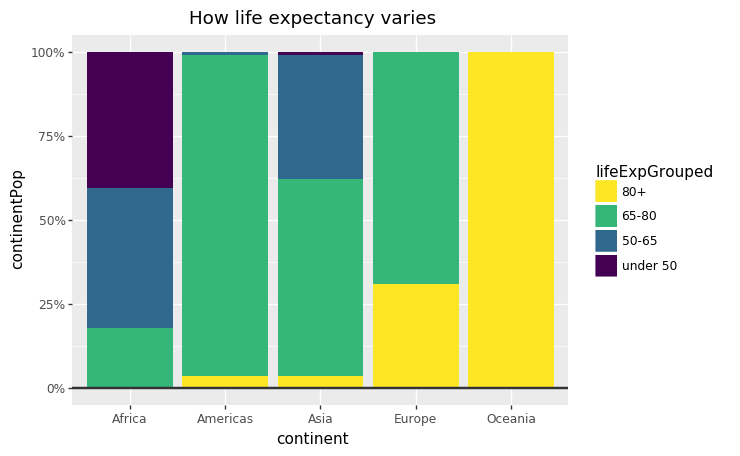
#Prepare data <- gapminder %>% filter (year == 2007 & continent == "Africa" ) %>% arrange (desc (lifeExp)) %>% head (5 )#Make plot <- ggplot (bar_df, aes (x = country, y = lifeExp)) + geom_bar (stat= "identity" , position= "identity" , fill= "#1380A1" ) + geom_hline (yintercept = 0 , size = 1 , colour= "#333333" ) + bbc_style () + labs (title= "Reunion is highest" ,subtitle = "Highest African life expectancy, 2007" )
## hide = gapminder.query(' year == 2007 & continent == "Africa" ' ).nlargest(5 , 'lifeExp' )
plotnine
= (ggplot(bar_df, aes(x= 'country' , y= 'lifeExp' )) + = "identity" ,= "identity" ,= "#1380A1" ) + = 0 , size= 1 , colour= "#333333" ) + # bbc_style() + = "Reunion is highest" ,= "Highest African life expectancy, 2007" ))
altair
= (alt.Chart(bar_df).mark_bar().encode(= 'country' ,= 'lifeExp' ,# color='country' = {'text' : 'Reunion is highest' ,'subtitle' : 'Highest African life expectancy, 2007' })
Make a stacked bar chart
Data preprocessing
## collapse-hide = (' year == 2007' )= lambda x: pd.cut('lifeExp' ],= [0 , 50 , 65 , 80 , 90 ],= ["under 50" , "50-65" , "65-80" , "80+" ]))'continent' , 'lifeExpGrouped' ], as_index= True )'pop' : 'sum' })= {'pop' : 'continentPop' })'lifeExpGrouped' ] = pd.Categorical(stacked_bar_df['lifeExpGrouped' ], ordered= True )6 )
0
Africa
under 50
376100713.0
1
Africa
50-65
386811458.0
2
Africa
65-80
166627521.0
3
Africa
80+
NaN
4
Americas
under 50
NaN
5
Americas
50-65
8502814.0
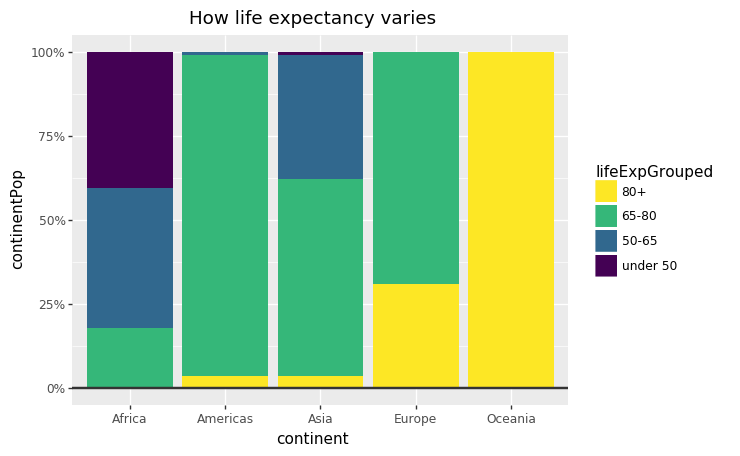
ggplot
#prepare data <- gapminder %>% filter (year == 2007 ) %>% mutate (lifeExpGrouped = cut (lifeExp, breaks = c (0 , 50 , 65 , 80 , 90 ),labels = c ("Under 50" , "50-65" , "65-80" , "80+" ))) %>% group_by (continent, lifeExpGrouped) %>% summarise (continentPop = sum (as.numeric (pop)))#set order of stacks by changing factor levels $ lifeExpGrouped = factor (stacked_df$ lifeExpGrouped, levels = rev (levels (stacked_df$ lifeExpGrouped)))#create plot <- ggplot (data = stacked_df, aes (x = continent,y = continentPop,fill = lifeExpGrouped)) + geom_bar (stat = "identity" , position = "fill" ) + bbc_style () + scale_y_continuous (labels = scales:: percent) + scale_fill_viridis_d (direction = - 1 ) + geom_hline (yintercept = 0 , size = 1 , colour = "#333333" ) + labs (title = "How life expectancy varies" ,subtitle = "% of population by life expectancy band, 2007" ) + theme (legend.position = "top" , legend.justification = "left" ) + guides (fill = guide_legend (reverse = TRUE ))
plotnine
# create plot = (= 'continent' ,= 'continentPop' ,= 'lifeExpGrouped' )+ = "identity" ,= "fill" ) + # bbc_style() + = lambda l: [" %d%% " % (v * 100 ) for v in l]) + =- 1 ) + # scale_fill_viridis_d = 0 , size= 1 , colour= "#333333" ) + = "How life expectancy varies" ,= " % o f population by life expectancy band, 2007" ) + = guide_legend(reverse= True )))
C:\Users\CHANNO.OOCLDM\AppData\Local\Continuum\anaconda3\lib\site-packages\plotnine\scales\scale.py:91: PlotnineWarning: scale_fill_cmap_d could not recognise parameter `direction`
warn(msg.format(self.__class__.__name__, k), PlotnineWarning)
C:\Users\CHANNO.OOCLDM\AppData\Local\Continuum\anaconda3\lib\site-packages\plotnine\layer.py:433: PlotnineWarning: position_stack : Removed 7 rows containing missing values.
data = self.position.setup_data(self.data, params)
# create plot = (= 'continent' ,= 'continentPop' ,= 'lifeExpGrouped' )+ = "identity" ,= "fill" ) + # bbc_style() + = lambda l: [" %d%% " % (v * 100 ) for v in l]) + =- 1 ) + # scale_fill_viridis_d = 0 , size= 1 , colour= "#333333" ) + = "How life expectancy varies" ,= " % o f population by life expectancy band, 2007" ) + = guide_legend(reverse= True )))
C:\Users\CHANNO.OOCLDM\AppData\Local\Continuum\anaconda3\lib\site-packages\plotnine\scales\scale.py:91: PlotnineWarning: scale_fill_cmap_d could not recognise parameter `direction`
warn(msg.format(self.__class__.__name__, k), PlotnineWarning)
C:\Users\CHANNO.OOCLDM\AppData\Local\Continuum\anaconda3\lib\site-packages\plotnine\layer.py:433: PlotnineWarning: position_stack : Removed 7 rows containing missing values.
data = self.position.setup_data(self.data, params)
altair
= (= 'continent' ,= alt.Y('continentPop' , stack= 'normalize' ,= alt.Axis(format = '%' )),= alt.Fill('lifeExpGrouped' , scale= alt.Scale(scheme= 'viridis' )))= {'text' : 'How life expectancy varies' ,'subtitle' : ' % o f population by life expectancy band, 2007' }= overlay = pd.DataFrame({'continentPop' : [0 ]})= alt.Chart(overlay).mark_rule(= '#333333' , strokeWidth= 2 ).encode(y= 'continentPop:Q' )+ hline
Make a grouped bar chart
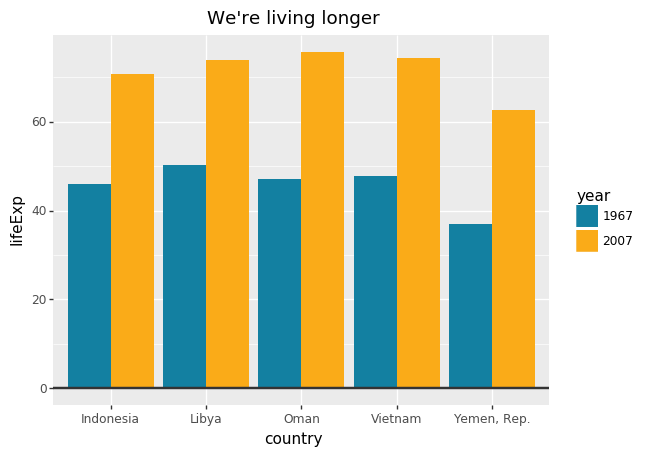
# hide = ('country' , 'year' , 'lifeExp' ' year == 1967 | year == 2007 ' )= ['country' ], columns= 'year' ,= 'lifeExp' )= lambda x: x[2007 ] - x[1967 ])5 , 'gap' )= [1967 , 2007 ],= ['country' , 'gap' ],= 'lifeExp' )
0
Oman
28.652
1967
46.988
1
Vietnam
26.411
1967
47.838
2
Yemen, Rep.
25.714
1967
36.984
3
Indonesia
24.686
1967
45.964
4
Libya
23.725
1967
50.227
5
Oman
28.652
2007
75.640
6
Vietnam
26.411
2007
74.249
7
Yemen, Rep.
25.714
2007
62.698
8
Indonesia
24.686
2007
70.650
9
Libya
23.725
2007
73.952
ggplot
#Prepare data <- gapminder %>% filter (year == 1967 | year == 2007 ) %>% select (country, year, lifeExp) %>% spread (year, lifeExp) %>% mutate (gap = ` 2007 ` - ` 1967 ` ) %>% arrange (desc (gap)) %>% head (5 ) %>% gather (key = year, value = lifeExp,- country,- gap) #Make plot <- ggplot (grouped_bar_df, aes (x = country, y = lifeExp, fill = as.factor (year))) + geom_bar (stat= "identity" , position= "dodge" ) + geom_hline (yintercept = 0 , size = 1 , colour= "#333333" ) + bbc_style () + scale_fill_manual (values = c ("#1380A1" , "#FAAB18" )) + labs (title= "We're living longer" ,subtitle = "Biggest life expectancy rise, 1967-2007" )
plotnine
# Make plot = (ggplot(grouped_bar_df,= 'country' ,= 'lifeExp' ,= 'year' )) + = "identity" , position= "dodge" ) + = 0 , size= 1 , colour= "#333333" ) + # bbc_style() + = ("#1380A1" , "#FAAB18" )) + = "We're living longer" ,= "Biggest life expectancy rise, 1967-2007" ))
altair
= (= 'year:N' ,= 'lifeExp' ,= alt.Color('year:N' , scale= alt.Scale(range = ["#1380A1" , "#FAAB18" ])),= 'country:N' )= {'text' : "We're living longe" ,'subtitle' : 'Biggest life expectancy rise, 1967-2007' }
Make changes to the legend
plotnine
# Remove the Legend + guides(colour= False )
+ theme(legend_position = "none" )
from plotnine import unit
ImportError: cannot import name 'unit' from 'plotnine' (C:\Users\CHANNO.OOCLDM\AppData\Local\Continuum\anaconda3\lib\site-packages\plotnine\__init__.py)
# Change position of the legend = multiline + theme(= element_line(color = "#333333" ),= 0.26 )
Work with small multiples
Do something else entirely
Make a dumbbell chart
# hide = ('country' , 'year' , 'lifeExp' ' year == 1967 | year == 2007 ' )= ['country' ], columns= 'year' ,= 'lifeExp' )= lambda x: x[2007 ] - x[1967 ])10 , 'gap' )
country
Oman
46.988
75.640
28.652
Vietnam
47.838
74.249
26.411
Yemen, Rep.
36.984
62.698
25.714
Indonesia
45.964
70.650
24.686
Libya
50.227
73.952
23.725
Gambia
35.857
59.448
23.591
Saudi Arabia
49.901
72.777
22.876
Nepal
41.472
63.785
22.313
Egypt
49.293
71.338
22.045
Tunisia
52.053
73.923
21.870
ggplot
#Prepare data <- gapminder %>% filter (year == 1967 | year == 2007 ) %>% select (country, year, lifeExp) %>% spread (year, lifeExp) %>% mutate (gap = ` 2007 ` - ` 1967 ` ) %>% arrange (desc (gap)) %>% head (10 )ggplot (hist_df, aes (lifeExp)) + geom_histogram (binwidth = 5 , colour = "white" , fill = "#1380A1" ) + geom_hline (yintercept = 0 , size = 1 , colour= "#333333" ) + bbc_style () + scale_x_continuous (limits = c (35 , 95 ),breaks = seq (40 , 90 , by = 10 ),labels = c ("40" , "50" , "60" , "70" , "80" , "90 years" )) + labs (title = "How life expectancy varies" ,subtitle = "Distribution of life expectancy in 2007" )
plotnine
Not available with plotnine.
altair
= (